Evaluation of RNA-based therapeutics’ efficacy
Team: Dr. Petros Avramopoulos, Reyhaneh Babaei, Dr. Claudia Klug, Dr. Bernhard Laggerbauer, Sabine Brummer, Anton Bomhard, Julia Auerswald
A widely employed approach to investigate microRNA function in vitro and in vivo are synthetic inhibitors (antimiRs) for these molecules. Although several chemical modifications have been invented, that warrant improved pharmacodynamics and stability against nucleases, their poor membrane penetrance, tissue- or cell-specificity of antimiRs are still significant problems that often obstruct progress toward therapeutic applications. The myocardium is one of the most challenging organs in this respect, but, on the other hand - as our work has shown - perhaps also one of the therapeutically most rewarding. Therefore, we aim to optimize the delivery and distribution of antimiRs against cardiac-specific miRNAs so that they show increased uptake, efficiency, and specificity in vivo. For this reason, we use different chemistries of antimiRs against specific miRNAs of interest in several models of heart failure. The distribution of fluorophore-coupled antimiRs is visualized by high-resolution microscopy, whereas its efficiency is determined by quantifying target mRNAs using deep RNA sequencing.
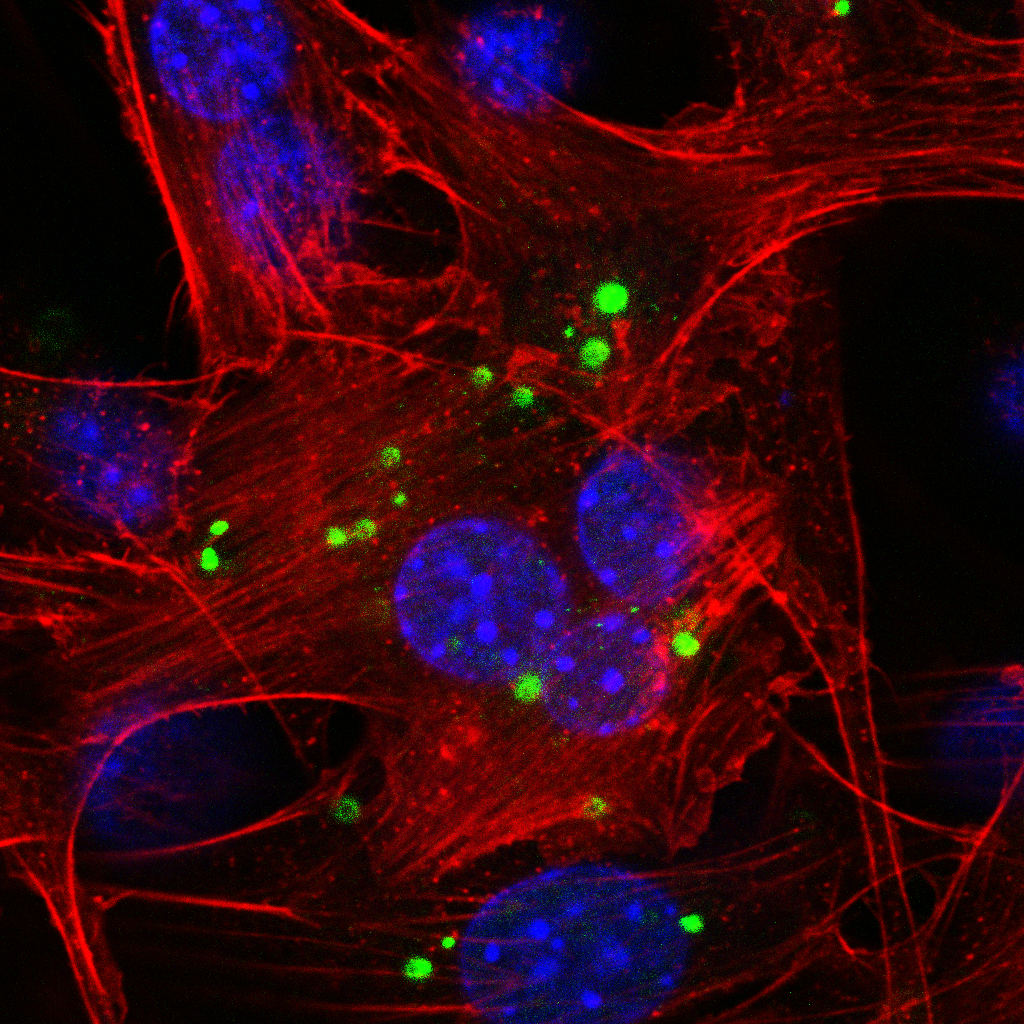
Confocal microscopy depicts the subcellular localization of a fluorescently-labeled microRNA inhibitor in mouse fibroblasts (NIH/3T3). Blue: DAPI staining, red: Alexa Fluor™ 647, green: 5'-FAM (6-fluorescein amidite (6-FAM))-labeled antimiR
The implication of microRNAs in cardiac disease
Team: Dr. Petros Avramopoulos, Reyhaneh Babaei, Dr. Dena Esfandyari, Gheo Idrissou, Dr. Anne Dueck, Dr. Claudia Klug, Dr. Bernhard Laggerbauer, Sabine Brummer, Anton Bomhard, Julia Auerswald
Although cardiac macrophages are known to exert critical functions during inflammatory responses, their specific tasks in cardiac disease are not yet fully understood. MicroRNAs, small, 20-24 nucleotide-long RNAs that negatively regulate the expression of target mRNAs, are already the focus of therapeutic approaches in humans. In our group, we study the role of microRNAs in a pressure overload and a myocardial infarction mouse model. We have already shown the role of several microRNAs of cardiac myocyte or fibroblast origin in the past. We now focus on the significantly deregulated microRNAs in the cardiac resident macrophage population.
To characterize the microRNAs of interest functionally, we employ a viral vector-based strategy for overexpression and genetic knockdown and chemically modified antimiRs for inhibition. Moreover, we have established the generation of functional microRNA knockouts using CRISPR in human induced pluripotent stem cells (hiPSC), which we then differentiate into macrophages. We then phenotype those microRNA knockout hiPSC-macrophages using assays such as phagocytosis, cell migration, and cytokine release assay. Deep RNA-seq and single-cell RNA-seq enable us to shed light on the molecular mechanisms responsible for the beneficial effects of microRNA manipulation in cardiac macrophages in vivo.
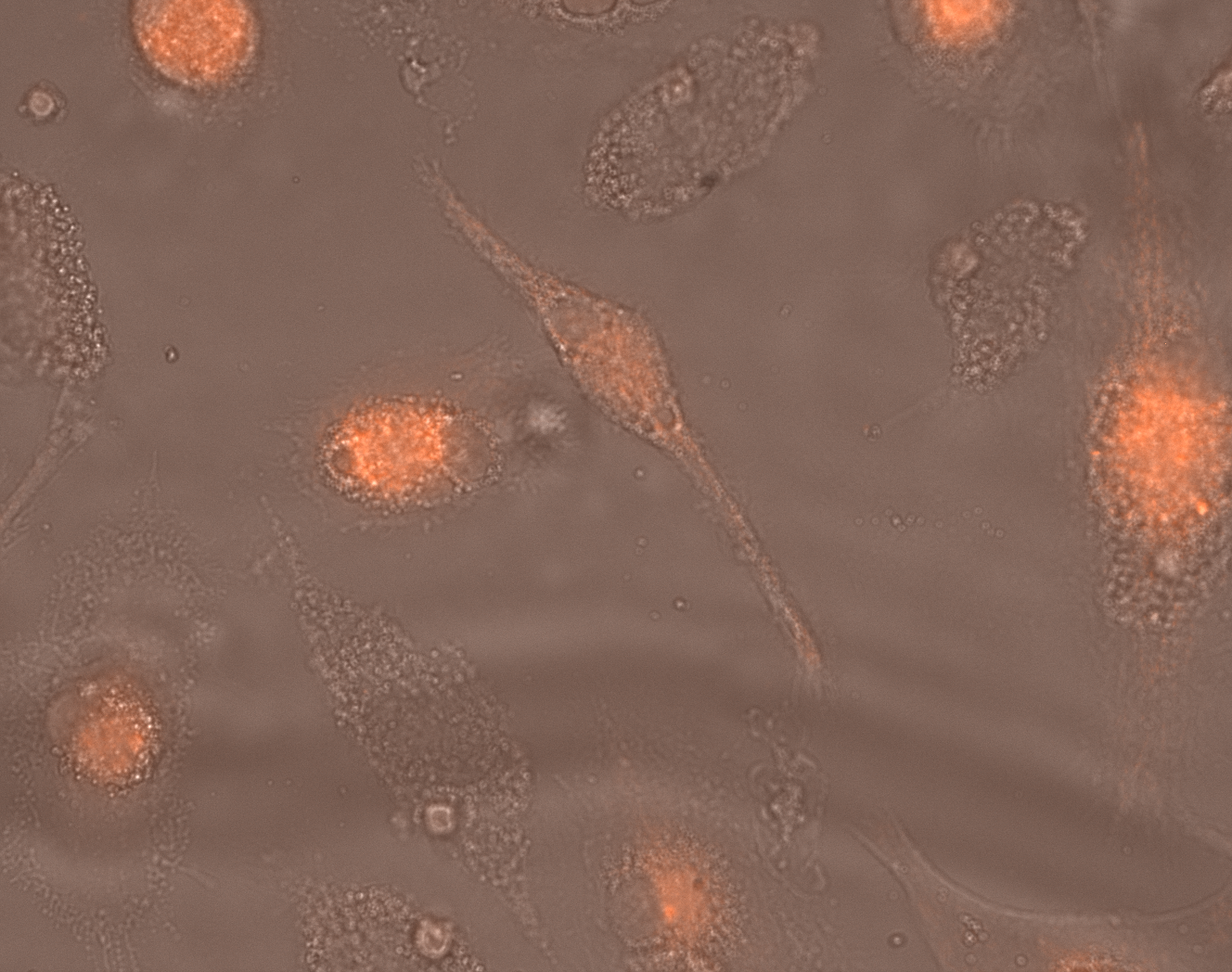
Time-lapse microscopy depicts a snapshot from hiPSC-macrophages during phagocytosis. Red: pHrodo™ Red – nonfluorescent at neutral pH, which only becomes fluorescent upon internalization

